
project page | paper | code

rg9360@princeton.edu
I am a PhD student at Princeton University, advised by Ben Eysenbach. Previously, I spent 1.5 years at Mila and Montreal robotics and AI lab. Before that, I completed my bachelors from NIT Nagpur. Broadly, my research goal is to develop simpler and scalable AI algorithms. I am interested in topics revolving reinforcement learning and probabilistic inference.
Please refer to Google Scholar for a complete list of my publications.

project page | paper | code
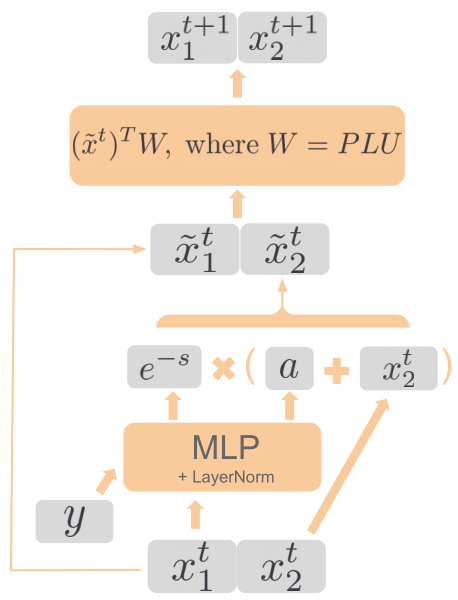
project page | paper | code
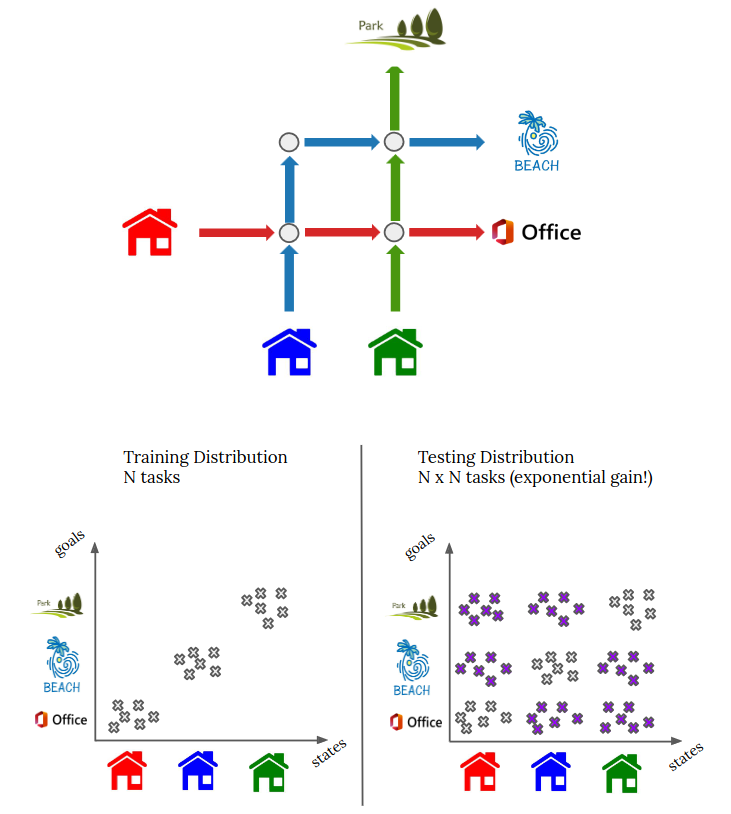
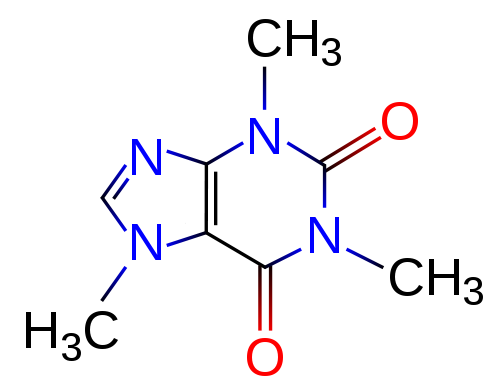
project page | paper | code
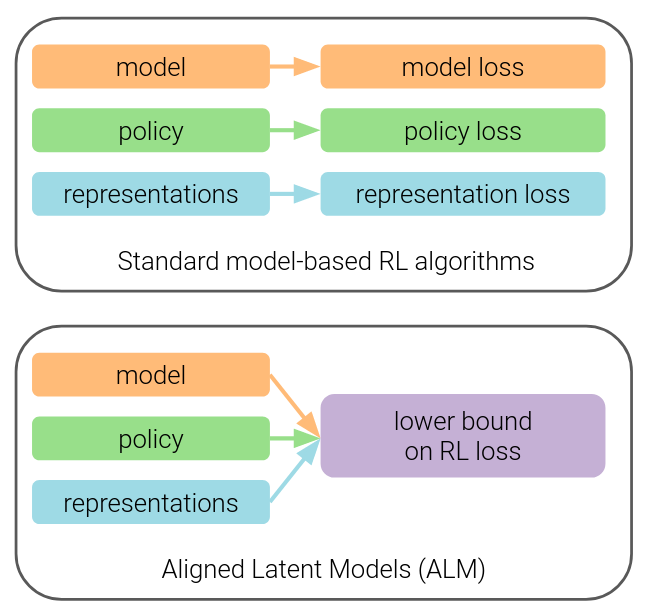
project page | paper | code
Last updated: October 2025.